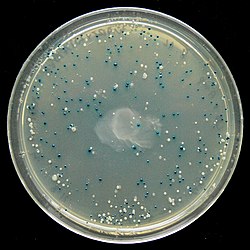
Blue white screening- Foriegn DNA and plasmid Dna are cut with restriction enzyme. Plasmid has gene for lactose hydrolysis lac z gene which codes for enzyme (β-galactosidase). Foriegn dna insert into lacz gene to form the recombinant. The recombinant dna is then transfered into a host cell , which are then grown in the presence of X-gal.
Result-
colonies with an insert-containing plasmid have a non-functional β-galactosidase, and remain the whitish-cream color of standard E. coli. On the other hand, intact β-galactosidase ( non recombinant plasmid )produces pigment from x-gal (included in the transformation plate medium), turning the bacterial colony blue.
Imagehttps://en.wikipedia.org/wiki/Blue_white_screen
Imagesourcehttp://www.slideshare.net/tejondaru/blue-white-selectionpresentation
Thank you for visiting my blog.Please feel free to share your comment on this article ,Please subscribe and share the articles to get more such articles.
Result-
colonies with an insert-containing plasmid have a non-functional β-galactosidase, and remain the whitish-cream color of standard E. coli. On the other hand, intact β-galactosidase ( non recombinant plasmid )produces pigment from x-gal (included in the transformation plate medium), turning the bacterial colony blue.

Imagehttps://en.wikipedia.org/wiki/Blue_white_screen
Imagesourcehttp://www.slideshare.net/tejondaru/blue-white-selectionpresentation
Thank you for visiting my blog.Please feel free to share your comment on this article ,Please subscribe and share the articles to get more such articles.